BACK TO RESEARCH WITH IMPACT: FNR HIGHLIGHTS
As the FNR marks 25 years since its creation, we highlight 25 examples of FNR-supported research with impact. Since returning to Luxembourg in 2010 as an FNR ATTRACT Fellow, Paul Wilmes has built an internationally leading research group focussed on unravelling the role that microbial communities – microbiomes – play in the environment, as well as in human health and disease.
Research into the human microbiome holds great promise for the development of novel diagnostic, prognostic and therapeutic approaches with expected far ranging impacts on human health and society. Microbial ecosystems are highly sensitive to changes in their environment, making them useful indicators for overall ecosystem health as well as linked human health. Once we understand these complex ecosystems, they may be harnessed for novel applications in sustainability and medicine.
“Microbiomes are central to human health as changes to the human microbiome are linked to a panoply of human diseases ranging from cancer to neurodegeneration. Microbiomes represent key systems of study as they directly link sustainability to ecosystem, environmental, and human health with the important potential for developing novel solutions for a sustainable and healthy future. Once we know how to rationally control microbiomes, they can be harnessed for new solutions including novel bioprocesses for energy production, the synthesis of sustainable materials, drugs or dietary regimens. ”Prof Dr Paul Wilmes Systems Ecology Group at the University of Luxembourg’s Luxembourg Centre for Systems Biomedicine and the Department of Life Sceinces and Medicine within the Faculty of Science, Technology and Medicine.
Paul Wilmes returned to his native country Luxembourg in 2010 from the University of Berkeley, thanks to an FNR ATTRACT Fellowship, which offers 5 years of funding up to 2 MEUR for promising young researchers to establish a research group in the country. Next to grants from various FNR funding instruments in the years that have followed, Wilmes also secured an ongoing ERC Consolidator Grant alongside funding from other European and international funding bodies.

Advances enabling deeper study of microbiomes
Over the past two decades, major advancements have taken place in the field of molecular microbial ecology, thanks to interdisciplinary approaches.
“Through the advent of molecular methods, we were for the first time able to probe deeply into the structure and function of complex microbial communities ranging from environmental to host-associated microbiota. This allowed us to study microbiomes without the need for traditional culturing techniques. This represented an essential paradigm shift as culture-based methods introduce important biases and lead to non-representative pictures of microbial diversity and function.”
The application of high-throughput DNA sequencing to microbiomes, also referred to as metagenomics, has revolutionised our understanding of the extent of microbial genomic diversity, especially in relation to the overall Tree of Life. This approach also paved the way for important discoveries such the use of CRISPR-Cas9 for gene editing, the subject of the 2020 Nobel Prize in Chemistry, a more complete understanding of biogeochemical cycles, and even the first genome sequence of SARS-CoV-2 through metatranscriptomics.
Pioneering metaproteomics
Wilmes played a founding role in the field of metaproteomics, the study of proteins expressed by microbiomes, in the first peer-reviewed publication from his PhD in 2004. Since this original publication, metaproteomics has since become an internationally growing field. Molecular microbial ecology has evolved significantly over the past 20 years, driven by rapid technological advancements and interdisciplinary collaborations.
“Thanks to a growing appreciation for the critical role of microbes in ecosystems coupled to technological advancements, the field of molecular microbial ecology has significantly evolved in the last two decades. Our research group and many others continue to explore novel approaches to unravel the complexities of microbial communities and their impact on environmental sustainability and human health. ”Prof Dr Paul Wilmes Systems Ecology Group at the University of Luxembourg’s Luxembourg Centre for Systems Biomedicine and the Department of Life Sceinces and Medicine within the Faculty of Science, Technology and Medicine.
Pioneering tools for molecular measurements over space and time
When first establishing the lab in the context of the ATTRACT programme, Wilmes’ research group pioneered wet- and dry-lab tools that enabled systematic molecular measurements of microbial consortia over space and time.
“The tools have allowed my group, in collaboration with others, to define the lifestyles of distinct microbial populations in the environment and to link these to their genetic and functional traits. Most recently, it has allowed us to predict the dynamics of microbiomes to a surprisingly high degree of determination. Such forecasting models will inform control strategies for microbial ecosystems which, in turn, will lead to new problem-directed applications for sustainable management practices such as integrated water reclamation and resource recovery from wastewater streams.”

The work of the Systems Ecology Group Wilmes has established revolves around microbiomes. The goal is to understand, predict, and control how they work, ranging from environment to human. The team is, for example, working to determine which roles they play in sustainable production processes or how they affect human health and disease.
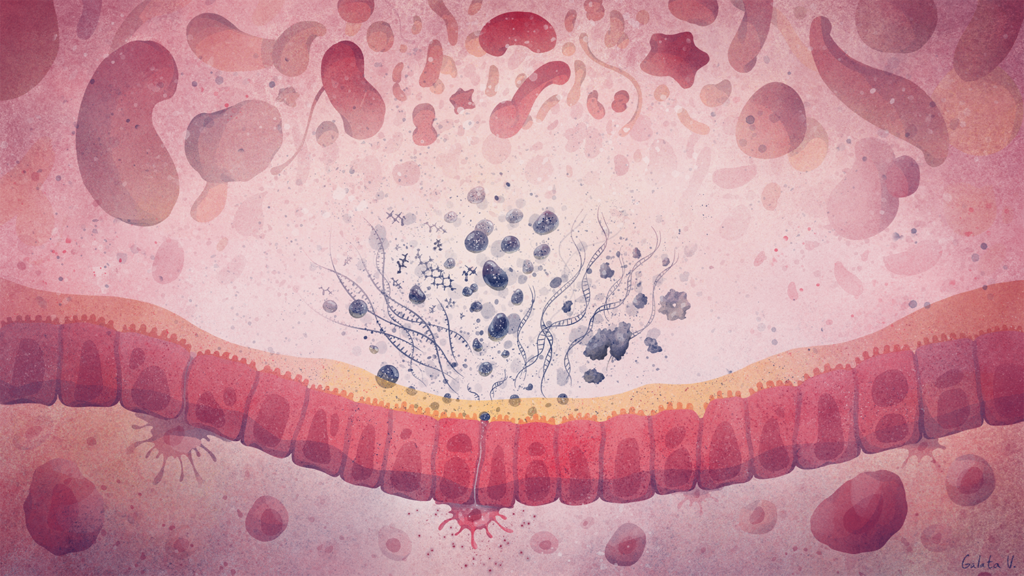
Using this methodology to study the human gut microbiome in diseases like Parkinson’s disease showed that the functions of the gut microbes are disrupted even if the overall structure of the microbiome appears the same. It also showed that when a child is born by vaginal birth, important immune system-stimulating bacteria pass from the mother to the baby, which could explain why babies delivered by caesarean are more prone to diseases linked to the immune system.
Read more about this research“The observation that overall microbiome-based functional differences are critical in the context of gut microbiome-linked, chronic diseases represents an essential paradigm shift in Biology and Medicine. These kinds of functional shifts may be triggered by factors which impact the gut microbiome, such as exposure to microorganisms or an unhealthy diet, which in turn may provide the ecological context for the development of diseases. ”Prof Dr Paul Wilmes Systems Ecology Group at the University of Luxembourg’s Luxembourg Centre for Systems Biomedicine and the Department of Life Sceinces and Medicine within the Faculty of Science, Technology and Medicine.
HuMiX and microGut: Organs-on-a-chip
Looking at the microbiome with a new software tool which the group has developed (PathoFact) revealed that certain genetic elements play particular roles in, for example, bacteria becoming resistant to antibiotics. This tool also helped show an increased infective competence in COVID-19. In order to better understand the human impacts of functional shifts in microbiomes, Wilmes’ group pioneered an “organ-on-a-chip” gut model called HuMiX alongside the expanded microGut system.
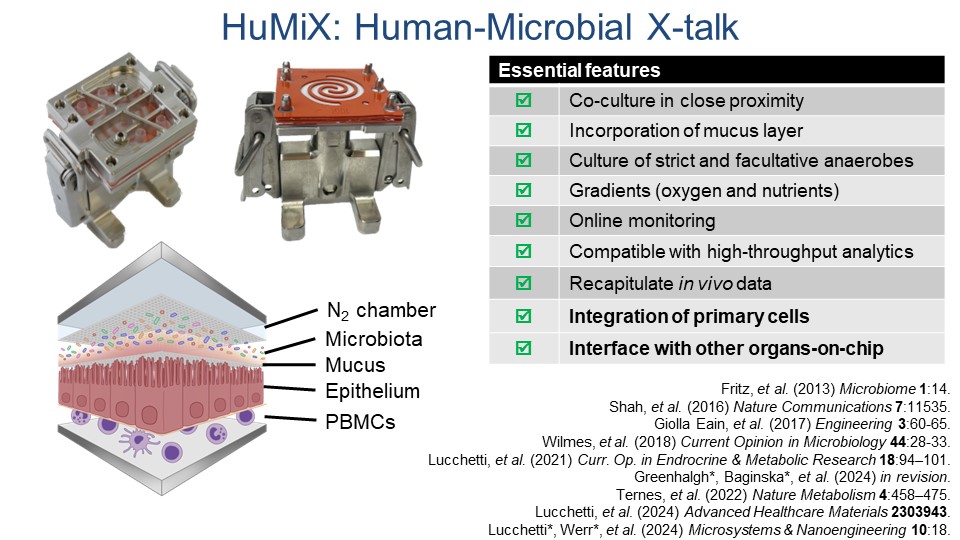
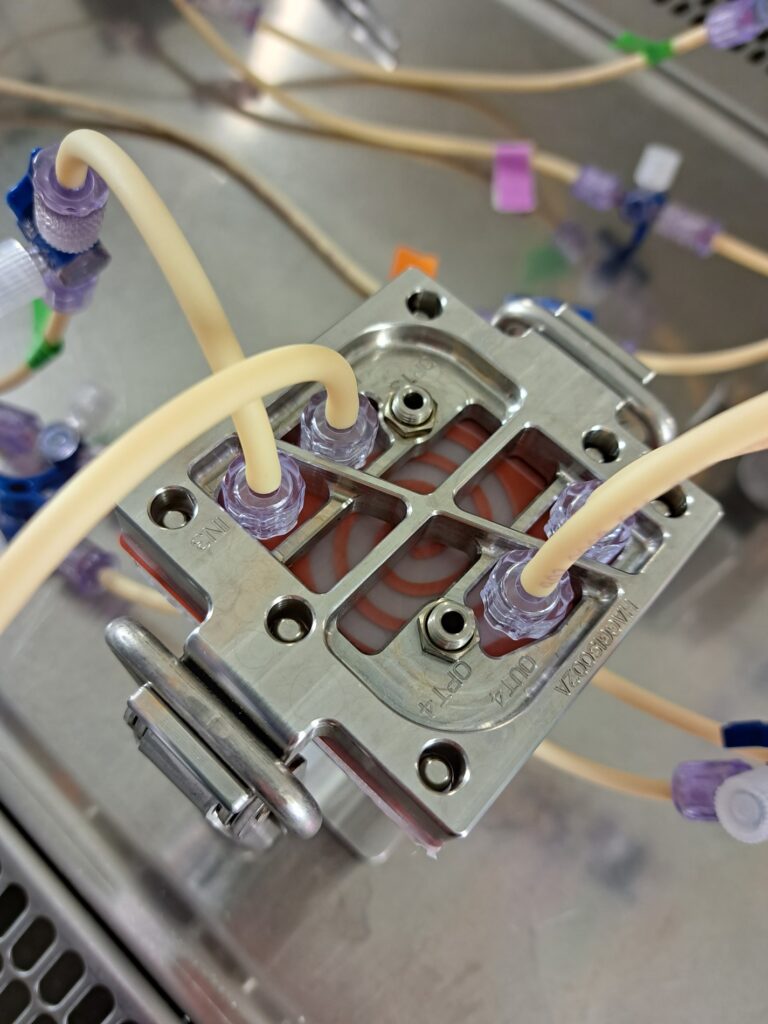

“HuMiX allows the co-culture of microbial and human cells under biomimetic conditions. It provides a platform for microbiome-focused mode of action studies against specific human disease backgrounds. It also allows for the in vitro study of essential characteristics which impact human health and disease such as the impact of diet and exposure to drugs on human physiology.”
“Our future research agenda will focus on understanding the molecules and molecular pathways by which the microbiome may trigger or exacerbate disease processes. This knowledge is essential for laying the foundation for establishing the microbiome as a therapeutic and preventative target. Once we have identified critical molecules and pathways, key research will be directed at how we might be able to steer microbiomes back to a balanced, healthy state. ”Prof Dr Paul Wilmes Systems Ecology Group at the University of Luxembourg’s Luxembourg Centre for Systems Biomedicine and the Department of Life Sceinces and Medicine within the Faculty of Science, Technology and Medicine.
Microbiome imbalances: Cause or effect of disease?
Microbiomes still hold much to be uncovered: In the context of the human microbiome, the major unanswered question is whether disease-linked microbiome shifts are a cause or an effect of disease. Researchers have established that there are imbalances in the microbiome in patients with, for example, cancer or neurodegenerative diseases, such as Parkinson’s Disease, but it remains to be determined whether these disturbances happen because of the disease, or if they could be a consequence of the disease occurring in the first place.

“All the findings highlighted here resulted from FNR-supported research projects. Four of our papers on the human microbiome were listed as milestones in the field by the journal Nature in 2019. It is fabulous to get such independent recognition for one’s own work and impact on a field of research. ”Prof Dr Paul Wilmes Systems Ecology Group at the University of Luxembourg’s Luxembourg Centre for Systems Biomedicine and the Department of Life Sceinces and Medicine within the Faculty of Science, Technology and Medicine.
In terms of microbiomes that underpin environmental systems, which are at the heart of sustainable resource management and a circular economy, Wilmes explains that rational control strategies will also have to be developed.
“We aim to unravel causal relationships and the linked mechanisms driving interactions within microbiomes and with their environment. Our field will evolve from being largely descriptive to proposing and implementing tangible solutions for human health and environmental sustainability. Through our wet- and dry-lab methodologies, including machine learning and AI, we aim to continue to play key roles in this exciting research and development field.”
Paul Wilmes on the impact of FNR funding
“The FNR has been instrumental in my growth as a scientist. During my PhD and postdoc in the UK, Germany and the US, I was able to benefit from publication and travel grants. When I aimed to become an independent scientist, I was able to achieve this through the ATTRACT programme of the FNR. Since then, the FNR has supported our research and development efforts through its various funding instruments, ranging from CORE to JUMP. Finally, the FNR has allowed us to build a strong research community in microbiology in Luxembourg with international radiance. This was in particular achievable through dedicated PRIDE and RESCOM funding.”
Paul Wilmes on training the next generation of scientists
“We aim to provide an open and rigorous training environment focused on community through which group members can subsequently pursue the careers of their choice whether within or outside of academia. My approach accounts for their different perspectives, skill sets and talents. The major impact on myself as a researcher is that I continue to learn a lot from group members. When they do well, such as by discovering something really new, it lifts us all up. These are the precious moments we all work so hard for.”
“Through FNR-supported research projects, I have successfully supervised 11 Master students, 15 doctoral students and 26 postdoctoral researchers. Dr Linda Wampach was the laureate of the 2019 FNR Award for Outstanding PhD Thesis. Dr Susana Martinez Arbas and Dr Laura de Nies received Excellent Thesis Awards from the University of Luxembourg in 2021 and 2022, respectively. Former members of our research group have gone onto successful careers of their own in academia and industry. Out of the former postdoctoral researchers, six became principal investigators immediately after leaving my laboratory [Prof. Nicolás Pinel, Prof. Mahesh Desai, Dr Emilie Muller (she was awarded a prestigious French CNRS research fellowship in 2016), Prof. Anna Heintz-Buschart, Dr Susheel Bhanu Busi, and Dr Laura de Nies (she was just awarded a prestigious Wellcome Trust Early-Career Award to conduct research at Oxford University in the UK)]. I aim to offer a diverse, equitable and inclusive research environment, in which group members openly share ideas and data, and in which they are also able to make mistakes. This policy also includes the publication of negative results (BMC Biology 2018, npj Parkinsons Dis 2024).”
Paul Wilmes on FNR memories
“When I was in the third and final year of my PhD in the UK, I applied for a grant under the FNR’s MA2 scheme to attend the major conference in my field at the time, focused on the dynamics of microbiomes in wastewater and organised by the International Water Association. The conference was held in Surfers’ Paradise on the Gold Coast in Australia. Once I had applied for the travel grant, I got a phone call from a somewhat unsettled FNR programme manager who wanted to make sure that I did not aim to benefit from FNR funding to go on sun-filled surfing vacation in Australia. I think I was able to reassure them. In any case, when I was later awarded the best paper prize at the conference, it was clear that all of this was legit. Suffice to say that I did get reimbursed for my travel to and accommodation in Surfers’ Paradise.”
| Project title | Funding instrument | Call year |
| Pathogenesis in the age of the microbiome | PRIDE | 2023 |
| Microbiome-Derived Molecules as Triggers of Disease | INTER Mobility | 2023 |
| Infective competence of the human microbiome in health and disease | CORE | 2023 |
| Towards a personalized approach to restore proteolytic homeostasis in digestive inflammation | INTER | 2023 |
| One Earth – Impacts and countermeasures to global change and effects on the biosphere | RESCOM | 2022 |
| COVID-19, vaccination and longer-term health consequences of COVID-19 in Luxembourg | CoVid19 funding instrument | 2021 |
| Multi-omics evaluation of microbial co-infection as marker of COVID-19 severity | CoVid19 funding instrument | 2020 |
| The dark metaproteome: identifying proteins of unknown function in the human gut microbiome | CORE | 2019 |
| 4th International Metaproteomics Symposium | RESCOM | 2019 |
| SIOtoSYS Lecture Series in Microbiology: From Single Organisms to Systems Ecology and Evolution | RESCOM | 2018 |
| Elucidating the factors governing microbial community composition | CORE | 2017 |
| iHuMiX – a personalised model of human gut for testing the efficacy of drugs and dietary compounds. | JUMP | 2017 |
| Microbiomes in One Health | PRIDE | 2017 |
| Non-invasive microbiome-derived multi-omic biomarkers for the early-stage detection and stratification of Parkinson’s disease | CORE | 2016 |
| Non-invasive microbiome-derived multi-omic biomarkers for early-stage colorectal cancer detection | CORE | 2015 |
| Linking environmental conditions to lifestyle strategies and to population-level genetic heterogeneity | CORE | 2015 |
| A microfluidics-based drug discovery platform emulating the human microbiome on chip | JUMP | 2015 |
| Systems Biology of Acidophile Biofilms for Efficient Metal Extraction | INTER | 2014 |
| Blood-borne microbiome-derived RNA biomarkers | JUMP | 2013 |
| Biomarkers for Alzheimer’s disease and Parkinson’s disease | CORE | 2012 |
| A microfluidics-based human-microbial cell co-culture device for resolving the molecular dynamics of syntrophy and antagonism in health and disease states | INTER | 2011 |
| Systems Biology of Natural Microbial Assemblages | ATTRACT | 2009 |
Publications related to this article
Pioneering metaproteomics: Wilmes & Bond, 2004, Environ Microbiol 6:911-20
Wet & dry lab tools:
- ISME J 2013, ISME Comm 2021a, Genome Biol 2016, Microbiome 2021
- Nat Comm 2014, ISME J 2015, Nat Comm 2020, Nat Microbiol 2021
- Nat Ecol Evol 2024
- Mov Disord 2017
- Nat Microbiol 2017, Microbiome 2022
- Front Microbiol 2017, Nat Comm 2018, ISME Comm 2021
- Trends Microbiol 2018, Cell Host Microbe 2022a,b
- Cell 2016, eLife 2019, Cell Rep 2023
New software tool:
- PathoFact; Microbiome 2021
- ISME Comm 2021a, Nat Comm 2022, eLife 2022
- Microbiome 2023 (Infective competence in COVID-19)
HuMiX:
- HuMiX; Nat Comm 2016
- Cell Rep 2019; Nat Metab 2022
- Adv Healthc Mater 2024
Related Funding Instruments
Related highlights
Spotlight on Young Researchers: The human gut microbiome and the clues it holds
Research is steadily painting a picture revealing the significance the human gut microbiome plays in health and disease. From gastrointestinal…
Read more
Mäin Element: Paul Wilmes
Read more
What microbes really do in our guts
Countless microorganisms live peacefully in our body, but they also can be involved in many diseases. To find out exactly…
Read more
Microbiome research: unlocking basic unknowns in human health
The number of genes possessed by the trillions of microbes in a human body outnumbers the host’s genes at least…
Read more
Diet and bacteria combination limits cancer progression
Researchers from the University of Luxembourg have discovered a combination of dietary factors and gut bacteria that inhibits the progression…
Read more
Study reveals caesarean birth impacts child’s immune system
When a child is born by vaginal birth, important immune system-stimulating bacteria pass from the mother to the baby, which…
Read more
Related highlights
25 examples of research with impact: The endless potential of plasma
As the FNR marks 25 years since its creation, we highlight 25 examples of FNR-supported research with impact. Over the…
Read more
25 examples of research with impact: A better understanding of rivers and their watersheds
As the FNR marks 25 years since its creation, we highlight 25 examples of FNR-supported research with impact. Starting with…
Read more
25 examples of research with impact: A geographical approach to a sustainable society
As the FNR marks 25 years since its creation, we highlight 25 examples of FNR-supported research with impact. One of…
Read more
25 examples of research with impact: Towards historical literacy
As the FNR marks 25 years since its creation, we highlight 25 examples of FNR-supported research with impact. Over the…
Read more
25 examples of research with impact: Securing online services
As the FNR marks 25 years since its creation, we highlight 25 examples of FNR-supported research with impact. In the…
Read more
25 examples of research with impact: Innovation beyond solid, liquid or gas
As the FNR marks 25 years since its creation, we highlight 25 examples of FNR-supported research with impact. Since arriving…
Read more